HVAC Digital Marketing Agency
Get a HVAC Marketing Agency Partner That Knows How Bring the Heat
The top 5% of HVAC contractors partner with Hook Agency for website design, search engine marketing, and paid ad management.
Contact Us Now


WEBSITE DESIGN
[Contact Us for Pricing] (Can be broken up)
We do custom website design with the Winning Website Formula. 100% Lead Generation focused.

SEARCH ENGINE OPTIMIZATION
[Contact Us for Pricing] Monthly
We help you rank higher on search with monthly reporting, content writing, link-building & technical edits.

PAID AD MANAGEMENT
[Contact Us for Pricing] Monthly
We can help manage your Google ad budget, keeping it tight and effective on a monthly basis.
We Want to Understand What Makes You Tick – And Be a Partner, Not Just a Vendor
We work with many home service businesses – and are dedicated to learning more about the HVAC industry regularly. We’ve driven over 10,000 qualified leads and can help you determine what people are searching for and get more website traffic and leads in your area. We not only do high-quality modern SEO for lead generation, we also make gorgeous websites and can maintain your HVAC website with taste, and our team of website designers and developers – alongside your team.
Did you know that HVAC businesses that invest in effective marketing strategies are 43% more likely to experience significant growth? Marketing plays a crucial role in the success of any business, and the HVAC industry is no exception. By leveraging the power of HVAC marketing services, you can propel your business forward and attract more customers than ever before.
HVAC marketing services encompass a range of strategies tailored specifically for home service providers like yourself. From digital advertising and search engine optimization (SEO) to social media management and customer reviews, these services help you establish a strong online presence, increase brand visibility, and drive more leads and conversions.
With the help of a reputable HVAC marketing company or agency, you can gain a competitive edge in your market. They become your trusted marketing partner, guiding you through the intricacies of reaching your target audience effectively. So why wait? It’s time to take advantage of HVAC marketing services and unlock the full potential of your business.
Challenges Faced by HVAC Businesses: Flat Sales, Rising Costs, and Low Brand Identity
Impact of Flat Sales on HVAC Businesses
Flat sales pose a significant challenge for HVAC companies. When customer demand remains stagnant, it becomes difficult to generate new customers and increase revenue. Without a steady stream of business, companies struggle to meet their financial goals and sustain growth. To overcome this hurdle, HVAC businesses need to explore innovative marketing strategies that target potential customers effectively.
How Rising Costs Affect Profitability in the Industry
Rising costs directly impact the profitability of HVAC businesses. As expenses such as fuel prices, equipment maintenance, and employee wages continue to climb, it becomes harder for companies to maintain healthy profit margins. To counteract this issue, businesses must find ways to optimize their operations and reduce costs without compromising on service quality. Investing in energy-efficient solutions or adopting smart technology can help streamline processes and minimize expenses.
Importance of Building a Strong Brand Identity to Stand Out From Competitors
Building brand awareness not only helps attract new customers but also fosters loyalty among existing ones. By creating a unique brand that resonates with customers’ needs and values, companies can differentiate themselves from competitors. This differentiation allows them to command higher prices for their services while maintaining customer satisfaction levels.
To enhance brand identity:
-
Consistently deliver exceptional service that exceeds customer expectations.
-
Utilize digital marketing strategies like search engine optimization (SEO), social media advertising, and content creation.
-
Develop a memorable logo and tagline that reflects the company’s values.
-
Engage with customers through various channels like email newsletters or online communities.
-
Showcase positive reviews or testimonials from satisfied clients.
By addressing these challenges head-on – flat sales, rising costs, and low brand identity – HVAC businesses can position themselves as top choices within the industry. With determination and strategic decision-making, they can create a solution-oriented approach, ensuring long-term success and growth.
The Power of HVAC Digital Marketing for Heating and Air Conditioning Businesses
Digital marketing has revolutionized the way HVAC businesses reach their target audience. With the advent of online platforms, such as social media and search engines, HVAC companies now have powerful tools to expand their reach and grow their business.
Reaching a Wider Audience for Your HVAC Business
By leveraging digital marketing strategies, HVAC businesses can tap into a vast online audience. With the right tactics, your business can gain visibility among potential customers who may not have known about your services. Through targeted advertising campaigns and search engine optimization (SEO) techniques, you can ensure that your business appears prominently in relevant online searches.
Cost-Effectiveness and Measurable Results Offered by Digital Marketing
One of the key advantages of digital marketing is its cost-effectiveness. Unlike traditional marketing methods, such as print ads or billboards, digital marketing allows you to allocate your budget more efficiently. Using platforms like social media advertising or content marketing, you can maximize your return on investment while reaching a larger audience.
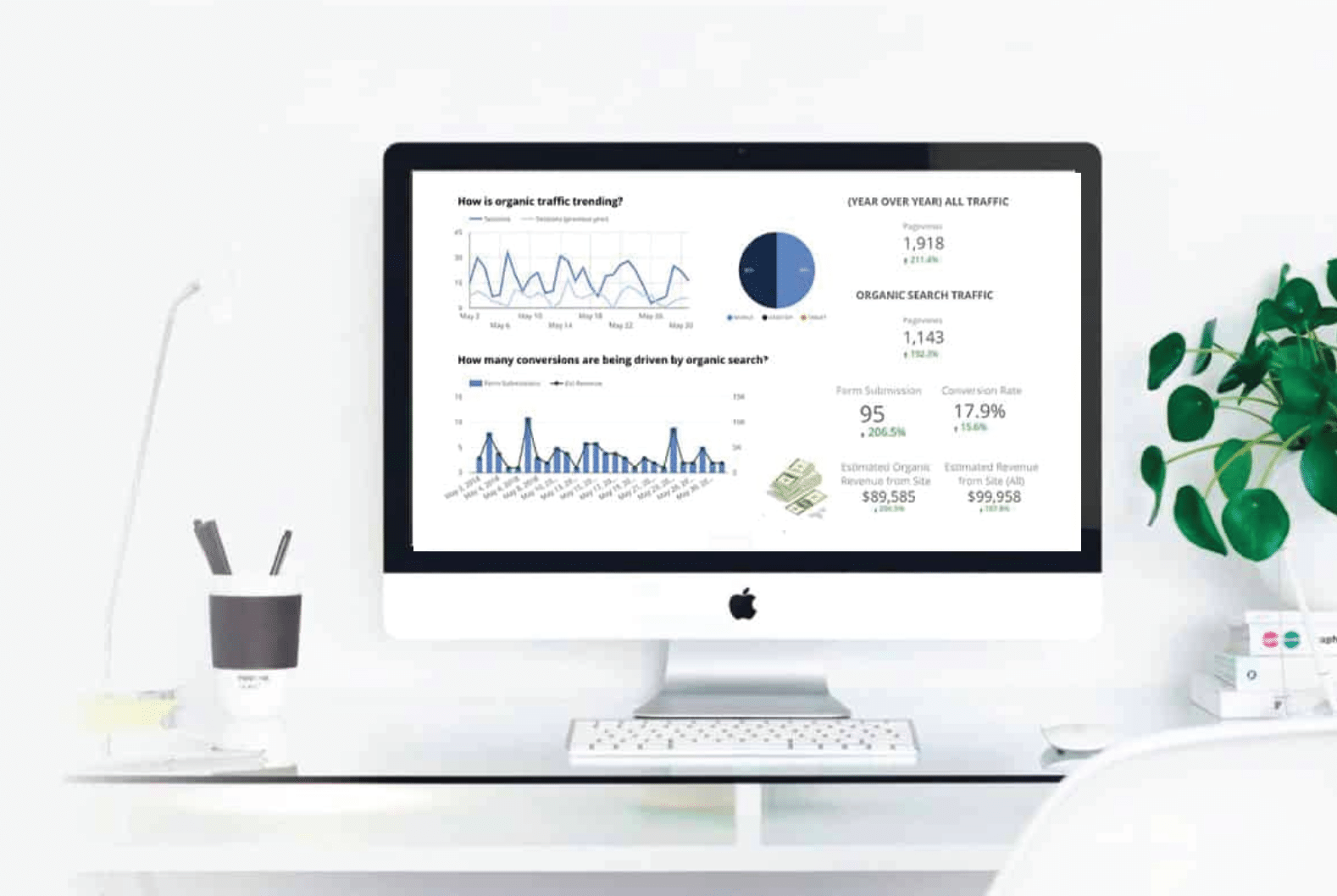
Moreover, digital marketing provides measurable results that allow you to track the success of your campaigns. Through analytics tools and data analysis, you can monitor website traffic, conversion rates, and customer engagement. This data-driven approach enables you to make informed decisions about where to allocate resources for maximum impact.
The Effectiveness of Digital Marketing for HVAC Companies
HVAC internet marketing has proven highly effective in boosting business growth within the industry. By partnering with a reputable digital marketing agency specializing in HVAC services, you can create tailored strategies that align with your goals. These agencies understand the nuances of the HVAC industry and can develop comprehensive digital marketing campaigns that drive leads and increase sales.
HVAC Digital Marketing Services: Boost Your Online Presence
Overview of Various Digital Marketing Strategies
To enhance your online presence as an HVAC business, it is crucial to explore different digital advertising strategies. By utilizing these techniques, you can effectively reach your target audience and promote your services. Consider the following options:
-
Website Design: Create an attractive and user-friendly website to engage visitors and encourage them to stay on your site. A well-designed website will keep customers interested in what you have to offer.
-
Search Engine Optimization (SEO): Improve website visibility on search engines by optimizing content and structure. This will help potential customers find you more easily when searching for HVAC services in their area.
-
Paid Ad Management: Efficiently manage paid advertising campaigns to reach a wider audience and drive targeted traffic to your website.
Effective HVAC Marketing Strategies for Heating and Air Conditioning Companies
Creating Engaging Content:
-
Educate customers about their HVAC needs through informative articles, blog posts, and videos.
-
Highlight the benefits of regular maintenance, energy-efficient systems, and indoor air quality.
-
Use storytelling techniques to engage readers and make the content relatable.
Importance of Local Optimization:
-
Optimize your website with local keywords to attract customers in your service area.
-
Create location-specific landing pages to improve visibility in search engine results.
-
Ensure your business information is accurate and consistent across online directories.
By implementing these HVAC marketing strategies, heating and air conditioning companies can effectively promote their services, attract new customers, and build long-lasting relationships. Whether creating engaging content that educates customers about their HVAC needs or utilizing email marketing campaigns to nurture leads, there are various ways to enhance your marketing efforts. Optimizing your website for local SEO can significantly increase visibility among potential customers in your service area.
Effective HVAC marketing involves understanding your target audience’s needs and providing valuable solutions. By delivering informative content through blogs, articles, and videos, you can position yourself as an industry expert while building trust with potential customers. Utilizing email marketing allows you to stay connected with leads by sharing useful tips and exclusive offers tailored to their interests. Lastly, optimizing for local SEO ensures that when people search for HVAC services in their area, they find your company at the top of the search results.
Maximizing Leads and Booked Jobs
HVAC businesses face numerous challenges, such as flat sales, rising costs, and low brand identity. However, by leveraging the power of HVAC digital marketing, heating and air conditioning companies can overcome these obstacles and achieve remarkable results. With effective HVAC marketing strategies, you can enhance your online presence, attract more leads, and ultimately increase your booked jobs.
So, how can you make the most out of your heating and air conditioning company marketing? By partnering with an experienced HVAC digital advertising agency that understands your industry. We will help you navigate the complex world of online marketing and develop tailored strategies to reach your target audience effectively. From search engine optimization (SEO) to social media advertising and content creation, they will implement a holistic approach to maximize your visibility in the digital landscape.
To take your HVAC business to new heights, it’s time to invest in comprehensive marketing solutions that drive results. Don’t let your competitors steal the spotlight—seize this opportunity to showcase your expertise and connect with potential customers. Get started today and watch as your leads soar and booked jobs multiply.
With our deep understanding of the HVAC industry, we can help you establish a strong online presence. Our ambition extends beyond enhancing your online visibility — we're committed to driving sustainable growth.
Our HVAC marketing & lead generation services are meant to aid your business, by being diligent about monthly reporting, providing a marketing dashboard, and taking these things off your plate:
- Our specialty contractor-focused writers + SEO Specialists will sit down with you to understand your audience and market and ask the right questions before diving into the data. (We also do competitive analysis and cross-section this with your ideas to find the money-making keywords.)
- We have relationships to 100’s of websites where we can get links from their website to yours – by writing useful articles on their site (one of the biggest ranking factors to get higher on Google.)
- We seek to understand your brand and your tone of voice and push to create value for your audience.
Our Process
- Quick intro call
- Strategy Meeting + Proposal
- Targeting Kickoff + We start work
- 1 month – Workshop + Revision
- Monthly reporting rhythm
Commonly Asked Questions
Why are you focused on the HVAC industry?
We love contractors!
And – we started in the roofing marketing/lead generation space, and have understood many of the same (marketing/lead generation) needs exist on the HVAC side. We currently have a couple of clients in the HVAC space, and love working with them – and so we are looking to help more HVAC contractors.
How long does it take for HVAC marketing strategies to show results?
The timeline for seeing results from HVAC marketing strategies can vary depending on factors such as competition level, budget allocation, and the specific tactics used. While some improvements may be noticeable within a few weeks or months, it generally takes several months for a comprehensive marketing campaign to yield significant results.
Can I handle my own HVAC digital marketing without professional assistance?
While it’s possible to manage some aspects of HVAC digital marketing on your own, partnering with professionals who specialize in this field can greatly enhance your chances of success. Digital marketing experts have the knowledge and experience needed to develop effective strategies tailored specifically for the HVAC industry while staying up-to-date with the latest trends and best practices.
How much should I budget for HVAC marketing services?
The budget for HVAC marketing services can vary depending on your specific goals, target audience, and the scope of the campaign. It’s important to consider factors such as website development, search engine optimization, content creation, social media advertising, and ongoing maintenance. Working with a professional agency will help you determine a suitable budget based on your unique requirements.
Can HVAC digital marketing help my business generate more leads?
Absolutely! HVAC digital marketing is designed to increase your online visibility and attract more qualified leads to your business. By optimizing your website for search engines, leveraging social media platforms, running targeted advertisements, and creating valuable content, you can effectively engage with potential customers and drive them toward booking your services.
What does your website design process look like?
We sit down with you and your leadership for our 4 step simple discovery. We identify your ideal customers, we walk through a positioning process, identify ‘trust badges’ and ‘trust factors’ for your site, and talk about what imagery will be persuasive to the people you serve.
What sets apart effective HVAC marketing strategies from ineffective ones?
Effective HVAC marketing strategies are those that take a comprehensive approach to reach and engage with your target audience. They involve utilizing various channels such as SEO, paid advertising, social media platforms, email marketing, and content creation to create a cohesive brand presence that resonates with potential customers. Ineffective strategies often lack consistency or fail to address the unique challenges faced by HVAC businesses in today’s competitive landscape.
What other ideas do you have for an HVAC company looking to drive leads?
The first couple moves for locally-focused contractors are Google My Business, Truck Wraps, and setting up your social media and posting about your company. Since we tend to work with growth-mode HVAC companies, the biggest moves you can make when you’re more mature are referral partnerships, and Google. Whether that’s paid ads, Google Guaranteed/local services, getting more reviews, and anything on the SEO side (which we really have mastered.) Beyond that it’s all about social proof – do you have testimonial videos? That’s a great next step for a lot of HVAC companies if they don’t have them yet.
Are You a Home Service Business Who Wants to Increase Your Qualified Leads?
Contact Us Now100+ 5-Stars

Award-Winning

Industry-Vetted