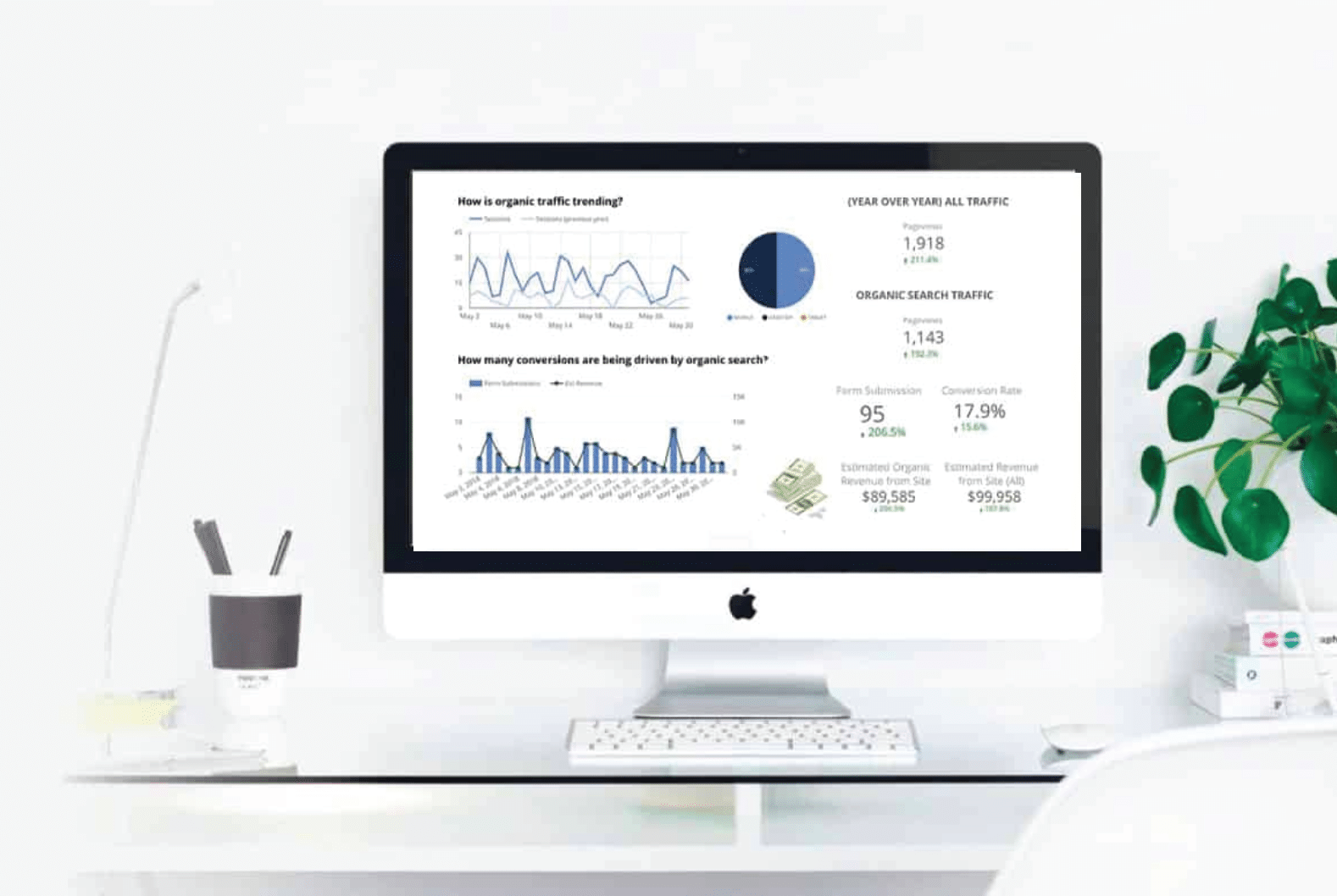
Get More Leads for your Home Services Business
Get higher on Google by partnering up with our results-driven home services SEO team. We know how to bring you more traffic and leads, ultimately allowing you to work the best jobs that you are looking for.
- We conduct competitive research into high-intent keywords that your ideal customers are searching for.
- We develop online campaigns with blog articles and create landing pages, which target those particular keywords.
- We get you high authority links so you become a trusted resource within your community.
100+ 5-Stars

Award-Winning

Industry-Vetted

Home Services SEO That Will Get you the Interested Leads You’re Looking For
You want to be intentional with your lead gen efforts. As much as you want to increase the number of leads you’re getting, you also want to make sure that you are receiving quality leads at the same time. Don’t waste your time with leads that never turn into paying customers.
By partnering up with a home services SEO company that specializes in contractors, you can ensure that your marketing spend doesn’t go to waste. Our expert team gets your ideal clients onto your site with our proven SEO process.
- We take the time to get to know your company and the ideal audience that you’re trying to target
- We do strategic research on the top money-making keywords and phrases in your industry
- We perform a detailed competitor analysis to see what terms your competition is ranking for and create content that will outrank them

Content
We can handle keyword targeting and writing user focused SEO driven content for you and your team.

Links
Backlinks are essentially votes to your website’s rankings. We earn highly authoritative relevant links back to your site from around the web.

Technical SEO
We identify all of the search terms that people are looking for across the web about your business and target your pages accordingly.
Monthly SEO + Reporting, and a Live Marketing Dashboard = Starting at [Contact Us for Pricing]/month
Contact Us Now
Fill in your information below and we’ll reach out the schedule a time to talk about your goals for your contractor company.
How long does it take to get results?
We generally say – 6-9 months for a clear upward trajectory on traffic or 9-12 months for obvious R.O.I.
How much does it cost?
Our SEO services aren’t the cheapest, but we know what works for home services + contractors. SEO is for people serious about turning their website into a traffic magnet. Contact us for pricing.
What makes Hook SEO services different?
We pride ourselves on being the ‘best SEO company in the world that ONLY does SEO for contractors.’ All of our systems are built around getting you more leads, and saving you time – with a business owners mentality.

Work With Home Services SEO to Get Ahead of Your Competition
Search engine optimization for home services is the process of improving your site and its brand reputation online so that it ranks highly in SERPs for keywords related to your home services. The higher you rank, the better your chances are of attracting online visitors and customers to your site.
It doesn’t matter how flashy or attractive your website is if you aren’t making efforts to direct quality traffic to it. If you have a website that isn’t getting high-quality conversions and leads, it’s going to be tough for you to get the results you want in order to help your business succeed. Here’s where we come in.
Our trained team of SEO specialists can help you get more people on your site, meaning you’ll have more opportunities to convert them into leads. Our proven strategy has helped a number of contractors just like you become authorities in their industry and level up their online presence.

We Are a Google Focused Agency
What does that mean?
You don’t have a bunch of Facebook ad guys, trying to do Google stuff as a side project.
Our main goal is to get you higher on Google and to make it easy for someone to contact you.
We don’t do RECRUITING MARKETING – although I know that’s a need.
We say “no” a lot so we’re the best at ONE THING.
- We say NO to non-construction non-home services marketing.
- We say no to marketing opportunities that aren’t exclusive to Google.
- We choose to only partner with high-quality, helpful home service contractors to provide superior service. We say no to asshole customers, so our writing team stays well-rested and eager to help.
Do you want to become the next remarkable Hook Agency customer?
Set up a time to chat with us, and we’d love to see if we can be of service to you. We’re ready to serve you, and will help you find a good fit for a contractor marketing agency, even if we’re not the right fit!
Why Home Services SEO is Essential in Today’s Digital Landscape
In today’s fast-paced digital age, people are increasingly turning to search engines for every need, including finding reliable home services. It’s no longer about just having a digital presence; it’s about ensuring that presence is optimized and positioned at the top of search results.
Why? Because 75% of searchers never go past the first page of Google.
If your home services business isn’t on that first page, you’re potentially missing out on a significant chunk of potential clients. That’s why SEO isn’t just an option; it’s a necessity for home service providers.
Our Home Services SEO strategies aren’t about just increasing the number of hits on your website but directing the right kind of traffic – users genuinely interested in what you have to offer. Whether you’re an HVAC, Plumbing, Roofing or other home service business!
So, when we talk about ‘quality traffic,’ we’re talking about real people, with real needs, that your business can genuinely satisfy.

Building Trust with Transparent Home Services SEO Reporting
At the heart of any successful partnership is trust. With many SEO companies, clients often feel left in the dark, unsure of where their money is going or what results they’re actually getting. That’s not how we operate.
We believe in full transparency. Our monthly reports provide clear insights into what we’re doing, how your keywords are performing, and the tangible results we’re delivering. This isn’t just about showcasing our success; it’s about building a relationship based on trust and accountability.
Our approach to Home Services SEO isn’t about quick wins or shortcuts.
It’s about creating lasting, organic results that not only elevate your online presence but build a sustainable brand reputation.
So, are you ready to light up the digital world with your home services? Dive deep into the realm of search engine optimization tailored for you, and let’s craft a success story together. Reach out today and embark on this transformative journey with us.
Our Process
- Quick intro call
- Strategy Meeting + Proposal
- Targeting Kickoff + We start work
- 1 month – Workshop + Revision
- Monthly reporting rhythm
Commonly Asked Questions
What is SEO?
Search engine optimization for home services is the process of improving your site and its reputation online so that it ranks highly in SERPs for keywords related to your home services. The higher you rank, the better your chances are of attracting online visitors and customers to your site.
Am I at a point where SEO is worth it and I should hire an agency?
This decision is entirely up to you. It does depend on your flexibility, motivation to learn, and the depth of your website Backed by your research, you could implement basic SEO strategies yourself. But you could also uncover a powerful strategy with the help of an expert.
Will I be locked in an SEO contract?
Absolutely not. But we talk about clear ROI in 6-9-12 months, and it starts to compound after that. We suggest people don’t work with an SEO company if they don’t plan on thinking long-term about SEO. We are probably not the best choice if you aren’t willing to work with us for a year or longer.
What kind of results should I anticipate from SEO?
You do not need to wait years to see results if you partner with Hook Agency. We build links in your website to trustworthy domains, which will help generate SEO results for you quickly. You can expect to notice profitable returns on your investment between 7-12 months.
What happens if I cancel SEO?
Almost everything we do will continue to benefit for a long time. Writing guest articles for other websites that link back to you, the content we write on your site, and in some cases, the P.R. mentions we’ve earned back to your site will continue to benefit you and potentially even increase lightly in effectiveness over the next 6-12 months after you stopped working with us / even if you stopped working with us. That being said NEW EFFORT regularly, is what will ensure that that graph doesn’t level off and even start dropping in the long-term.
What separates you from other cheaper SEO providers?
We combine a unique style of effective keyword targeting and brand identity, which attracts qualified prospects for you. The team’s conversations, actions, and motivations are centered around increasing revenue and customers for you.
Articles Related to Home Services SEO Marketing
All ArticlesAre You a Home Service Business Who Wants to Increase Your Qualified Leads?
Contact Us Now100+ 5-Stars

Award-Winning

Industry-Vetted